Evolutionary Optimization Module
Last modified: September 16, 2022
Contents
Introduction
Feature Selection, Weighting and Genetic Algorithms
Feature selection is the technique of selecting a subset of good features out of a larger featureset to obtain the features which are suitable for the best possible classification. Feature weighting is a generalization of feature selection with real-valued weights between [0,1] instead of the binary values 0/1.
Due to the fact that feature selection and weighting are NP-hard optimization problems, a complete and correct solution is not easy to determine. One way to get an approximate solution is by using genetic algorithms. Even though feature weighting is a generalization of feature selection and should thus be able to produce better classification results, practice has shown that feature selection produces better recognition rates in most cases. It is thus recommended to use feature selection rather than feature weighting [Bolten2012].
Genetic algorithms, proposed by Holland, are adaptive problem solvers that find solutions based on the mechanics of natural selection and natural genetics. For more information, see a genetic algorithms text such as [Holland1975]
How the optimization can be used after training the classifier is described in the training tutorial.
Usage
There are two ways to use the optimization:
- Graphical User Interface: Biollante
- Python Script API
These two approaches are equivalent, although due to the required runtime the script API is recommended.
User Interface Biollante
"Biollante is the result of an experiment in genetic engineering. The creature was created by splicing together the DNA of three organisms: a rose, a human female (Erica, daughter of the scientist), and Godzilla himself."
For our purposes, Biollante is a GUI for optimizing kNN classifiers using genetic algorithms. A genetic algorithm is used to adjust the selection and weight of each feature.
Overview
To use Biollante from the Gamera console window select File -> Biollante. From the Biollante window select File -> Open from XML to open a Gamera XML file containing training data or load with File -> Open existing classifier an existing classifier from the gamera main window. Once the training data has been loaded you can start and stop the optimization process. After the optimizer has finished, the results can be saved using File -> Save to XML or File -> Save to XML as.
Classifier specific settings are configurable through the Classifier menu. With Classifier -> Copy classifier it is possible to create a copy from the loaded classifier for the optimization process. The menu entry Classifier -> Classifier to Gamera-GUI allows the usage of the optimized classifier in the Gamera main window. Through Classifier -> Load settings from XML a settings xml file will be loaded and applied.
The used feature set can be modified with Classifier -> Change feature set. With the menu entries Classifier -> Reset selections and Classifier -> Reset weights it is possible to discard already chosen optimizations.
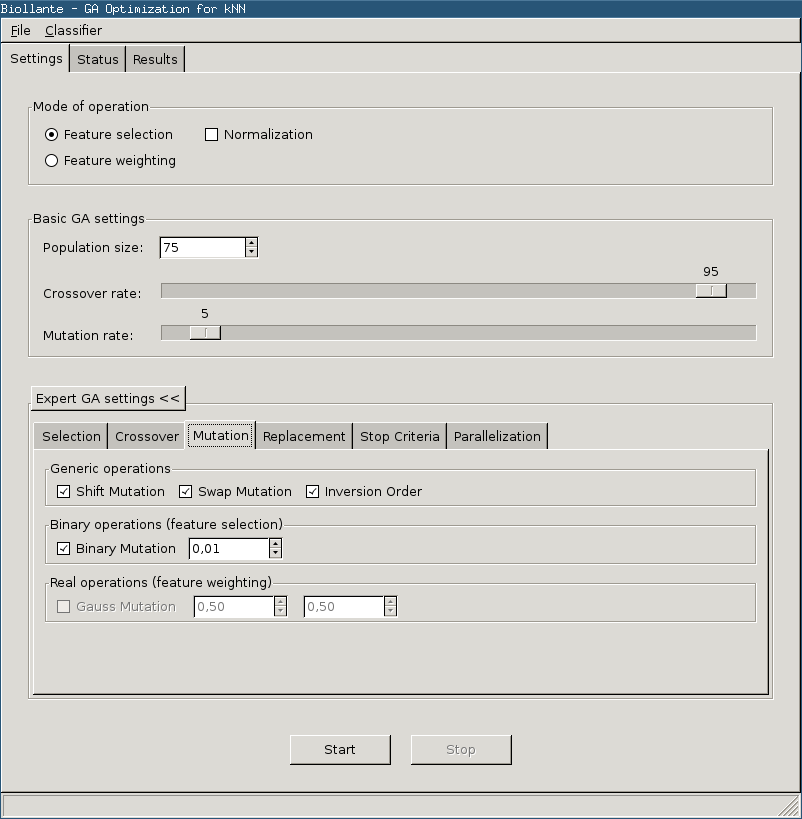
Settings Page
The Biollante settings page offers the control over the evolutionary optimization process. It is possible to select which kind of optimization is applied: selection or weighting.
Additionally, the basic parameters population size, crossover rate and mutation rate can be adjusted. In the optional available Expert GA settings it is possible to select and fine-tune each used genetic algorithms operator (for further information see below). These settings are predefined with useful default values.
The crossover rate and mutation rate are expressed in percentages.
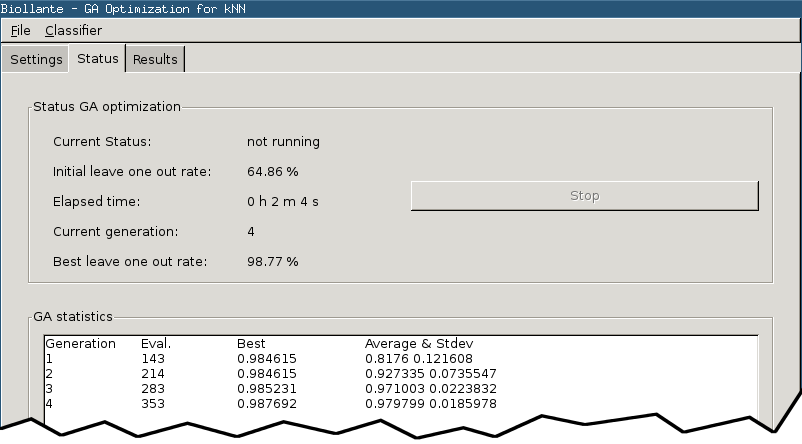
Status Page
The Biollante status page displays information about the current state of the optimization and allows to stop the process manually.
- Current Status
- Flag which indicates whether the optimization is currently running or not.
- Initial leave one out rate
- The initial leave-one-out performance of the classifier.
- Elapsed time
- Clock which measures how much time elapsed since optimization start.
- Current generation
- The current number of generations (of the genetic algorithm).
- Best leave one out rate
- The best leave-one-out performance achieved so far.
- GA statistics
- Text field which shows statistical information about the optimization process like the number of fitness evaluations, average and stdev of fitness values within each generation.
Result Page
The result page graphically displays the best selections or weights in a "bar graph" style.
Script Usage
The optimization module is also fully operable from Python directly.
The following listing clarifies the usage of the optimization module. All functions are explained in the function reference.
from gamera.core import *
init_gamera()
from gamera import knn, knnga
if __name__ == "__main__":
# Load the trainingdata and create a classifier object
classifier = knn.kNNNonInteractive("classifiers/traindata.xml",
features = 'all',
normalize = False)
# Create and configure the GA objects and settings
baseSettings = knnga.GABaseSetting()
baseSettings.opMode = knnga.GA_SELECTION
baseSettings.popSize = 75
baseSettings.crossRate = 0.95
baseSettings.mutRate = 0.05
selection = knnga.GASelection()
selection.setRoulettWheelScaled(2.0)
crossover = knnga.GACrossover()
crossover.setUniformCrossover(0.5)
mutation = knnga.GAMutation()
mutation.setSwapMutation()
mutation.setBinaryMutation(0.5, False)
replacement = knnga.GAReplacement()
replacement.setSSGAdetTournament(3)
stop = knnga.GAStopCriteria()
stop.setSteadyStateStop(40,10)
parallel = knnga.GAParallelization()
parallel.mode = True
parallel.thredNum = 4
# Combine each setting object into one main object
ga = knnga.GAOptimization(classifier, baseSettings, selection,
crossover, mutation, replacement,
stop, parallel)
# Run the optimization
ga.startCalculation()
# Lets have a look on the reported optimization progress and the results
print "Generation statistics", ga.monitorString
print "Selections:", classifier.get_selections_by_features()
It is recommended to use the settings from the Biollante GUI.
Function Reference
The optimization module follows a highly object oriented design.
Capsle Object GAOptimization
GAOptimization
GAOptimization (KnnObject classifier, GABaseSetting base, GASelection selection, GACrossover crossover, GAMutation mutation, GAReplacement replace, GAStopCriteria stop, GAParallelization parallel)
The GAOptimization constructor combines all GA-setting objects and a kNN-Classifier into one executable genetic algorithm which allows the optimization.
- KnnObject classifier
- a trained instance of a gamera kNN-classifier (no type checking is performed)
- GABaseSetting base
- a configured instance of the GABaseSetting class
- GASelection selection
- a configured instance of the GASelection class
- GACrossover crossover
- a configured instance of the GACrossover class
- GAMutation mutation
- a configured instance of the GAMutation class
- GAReplacement replace
- a configured instance of the GAReplacement class
- GAStopCriteria stop
- a configured instance of the GAStopCriteria class
- GAParallelization parallel
- a configured instance of the GAParallelization class
Note
No type checking is performed for the classifier object!
Getter
status
flag which indicates whether the optimization is in progress (True) or not (False)
generation
the number of the latest computed generation
bestFitness
the best fitness value which has occurred in the optimization progress so far
monitorString
string which contains some statistical information about the optimization process, like number of fitness evaluations, average and stdev of fitness values within each generation.
bestIndiString
string coded version from the best individual of each generation
Functions
startCalculation
startCalculation ()
Starts the evolutionary optimization progress
Note
After starting the optimization no changes in the used classifier object are allowed until the optimization is finished!
stopCalculation
stopCalculation ()
Manual stops the evolutionary optimization progress without respect to the stoping-criterias.
Note
This function blocks until the current generation of the optimization is finished. As a result it could take a long time until this functions returns!
Base Settings
GABaseSetting
GABaseSetting (opMode = GA_SELECTION, int popSize = 75, double crossRate = 0.95, double mutRate = 0.05)
The GABaseSetting constructor creates a new settings object with the basic parameters for GA-optimization for the later usage in a GAOptimization-object.
- opMode (optional)
- set optimization mode to GA_SELECTION or GA_WEIGHTING
- int popSize (optional)
- population size which is used in the GA-progress
- double crossRate (optional)
- crossover probability which is used in the GA-progress
- double mutRate (optional)
- mutation probability which is used in the GA-progress
Getter and Setter
opMode
flag which determines the mode of operation (can be set to the defined constants GA_SELECTION (0) or GA_WEIGHTING (1))
popSize
the population size
crossRate
the crossover probability (should be between 0.0 and 1.0)
mutRate
the mutation probability (should be between 0.0 and 1.0)
Individuals Selection Settings
GASelection
GASelection ()
The GASelection constructor creates a new settings object for the GA-optimization which specifies the used individuals selection method. This object can later be used in an GAOptimization-object.
Only one selection method can be chosen. Multiple settings will override each other.
Functions
setRoulettWheel
setRoulettWheel ()
Set the roulette wheel selection method for the GA-progress. This method picks up an individual proportional to the stored fitness value.
setRoulettWheelScaled
setRoulettWheelScaled (double preasure = 2.0)
Set the roulette wheel selection with linear scaled fitness values as selection method for the GA-progress.
- double preasure (optional)
- the selective pressure, should be in [1,2]
setStochUniSampling
setStochUniSampling ()
Set the stochastic universal sampling method as selection method for the GA-progress.
This method selects an individual proportional to the stores fitness value. Compared with the roulette wheel selection method all individuals are selected in only one step.
setRankSelection
setRankSelection (double preasure = 2.0, double exponent = 1.0)
Set the rank selection method as selection method for the GA-progress.
This method picks up an individual by roulette wheel based on his rank in the current population.
- double preasure (optional)
- the selective pressure, should be in [1,2]
- double exponent (optional)
- exponent (1 == linear)
setTournamentSelection
setTournamentSelection (int tSize = 3)
Set a deterministic tournament selection method for selecting the individuals in the genetic algorithm.
- int tSize (optional)
- the number of individuals in the tournament
Crossover Settings
GACrossover
GACrossover ()
The GACrossover constructor creates a new settings object for the GA-optimization which specifies the selected crossover-operators. This object can later be used in an GAOptimization-object.
Combination from different crossover-operators are possible. Each selected operator can later used with an equal probability within the optimization.
Functions
setNPointCrossover
setNPointCrossover (int n = 1)
Enables the classic n-Point crossover as recombination operator in the GA-optimization.
- int n (optional)
- the number of crossover points
setUniformCrossover
setUniformCrossover (double preference = 0.5)
Enables the classic uniform crossover operator in the recombination of the GA.
- double preference (optional)
- crossover-probability for each allele (should be in [0,1])
setSBXcrossover
setSBXcrossover (int numFeatures, double min, double max, double eta = 1.0)
Enables the simulated binary crossover operator for the real value encoded individuals used in feature weighting. For more details on this operator see: Deb, K. & Agrawal, R. B. (1995) Simulated Binary Crossover for Continuous Search Space. Complex Systems, 9, pp. 115-148.
- int numFeatures
- the number of used features. Usually this value is set to classifier.num_features.
- double min (optional)
- the minimum value which is allowed in an allele (usually 0.0)
- double max (optional)
- the maximum value which is allowed in an allele (usually 1.0)
- double eta (optional)
- the amount of exploration OUTSIDE the parents as in BLX-alpha notation
setSegmentCrossover
setSegmentCrossover (int numFeatures, double min, double max, double alpha = 0.0)
Enables the segment crossover operator for the real value encoded individuals used in feature weighting. This operator is a variation from the classical arithmetic recombination which performs a uniform choice in segment (arithmetical with same value along all coordinates).
- int numFeatures
- the number of used features. Usually this value is set to classifier.num_features.
- double min (optional)
- the minimum value which is allowed in an allele (usually 0.0)
- double max (optional)
- the maximum value which is allowed in an allele (usually 1.0)
- double alpha (optional)
- the amount of exploration OUTSIDE the parents as in BLX-alpha notation
setHypercubeCrossover
setHypercubeCrossover (int numFeatures, double min, double max, double alpha = 0.0)
Enables the hypercube crossover operator for the real value encoded individuals used in feature weighting. This operator is a variation from the classicial arithmetic recombination which performs a uniform choice in hypercube (arithmetical with different values for each coordinate).
- int numFeatures
- the number of used features. Usually this value is set to classifier.num_features.
- double min (optional)
- the minimum value which is allowed in an allele (usually 0.0)
- double max (optional)
- the maximum value which is allowed in an allele (usually 1.0)
- double alpha (optional)
- the amount of exploration OUTSIDE the parents as in BLX-alpha notation
Mutation
GAMutation
GAMutation ()
The GAMutation constructor creates a new settings object for the GA-optimization which specifies the selected mutation-operators. This object can later be used in an GAOptimization-object.
Combinations of different mutations-operators are possible. Each selected operator is used with an equal probability within the optimization.
Note
Only mutation-operators from the corresponding category (feature selection or weighting) are used later in the optimization based on the selection in the GABaseSetting-object.
Functions
setShiftMutation
setShiftMutation ()
Enables the shift mutation on the chromosome as mutation operator in the genetic algorithm. This means the allele between two points are right shifted.
setSwapMutation
setSwapMutation ()
Enables the swap mutation on the chromosome as muation operator in the genetic algorithm. This means two allele within the chromosome are exchanged.
setInversionMutation
setInversionMutation ()
Enables the inversion mutation on the chromosome as mutation operator in the genetic algorithm. This means the ordering from the allels between two points will be reversed.
setBinaryMutation
setBinaryMutation (double rate = 0.05, bool normalize = False)
Enables the classic bit-flip mutation for bitstring individuals. This means that this operator only effects feature selection.
- double rate (optional)
- the probability for mutation of an allele (should be in [0,1])
- bool normalize (optional)
- if true rate/chromosomeSize is used
setGaussMutation
setGaussMutation (int numFeatures, double min, double max, double sigma, double rate)
Enables the mutation based on a Gaussian distribution for real value individuals. This means that his operator only effects feature weighting. A random number is added to the mutated allele.
- int numFeatures
- the number of used features. Usually this value is set to classifier.num_features.
- double min
- the minimum value which is allowed in an allele (usually 0.0)
- double max
- the maximum value which is allowed in an allele (usually 1.0)
- double sigma
- the standard deviation of the gaussian distribution. This parameter determines the strength of the mutation.
- double rate
- the probability for mutating an allele (should be in [0,1])
Replacement
GAReplacement
GAReplacement ()
The GAReplacement constructor creates a new settings object for the GA-optimization which specifies the used replacement method. This object can later be used in an GAOptimization-object.
Only one replacement method can be chosen. Multiple settings will override each other.
Functions
setGenerationalReplacement
setGenerationalReplacement ()
Sets the generational replacement as replacement method for the GA-progress. This means that the new individuals in the intermediate population will replace the whole population.
setSSGAworse
setSSGAworse ()
Sets the steady state worse replacement as replacement method in the GA-progress. This means that the worst individuals are continuous replaced by better ones.
setSSGAdetTournament
setSSGAdetTournament (int tSize = 3)
Sets an steady state deterministic tournament as replacement method in the GA-progress. This means that the loosers of this tournament will be replaced in the following population.
- int tSize (optional)
- the number of individuals in the tournament
Stop Criteria
GAStopCriteria
GAStopCriteria ()
The GAStopCriteria constructor creates a new settings object for the GA-optimization which specified the termination condition of the optimization progress. This object can later be used in an GAOptimization-object.
A combination of different stop-criteria methods is possible. The first condition which becomes True will end the optimization.
Functions
setBestFitnessStop
setBestFitnessStop (double optimum = 1.0)
Enables the termination by attainment of the specified fitness rating measured by one individual.
- double optimum (optional)
- fitness rating
setMaxGenerations
setMaxGenerations (int n = 100)
Enables the termination by attainment of the specified number of maximal generations within the optimization process.
- int n (optional)
- the number of maximal generations
setMaxFitnessEvals
setMaxFitnessEvals (int n = 5000)
Enables the termination by attainment of the specified amount of fitness evaulations within the optimization process
- int n (optional)
- the number of fitness evaluations
setSteadyStateStop
setSteadyStateStop (int minGens = 40, int noChangeGens = 10)
Enables the termination after noChangeGens generations without improvements within the optimization after at least minGens generations.
- int minGens (optional)
- the minimum number of generations to be computed
- int noChangeGens (optional)
- the number of generations without improvements
Parallelization
GAParallelization
GAParallelization (mode = True, threads = 2)
The GAParallelization constructor creates a new settings object which controls the parallelization within the GA-progress. It is used later within an GAOptimization-object.
- mode (optional)
- enable (True) or disable (Flase) the parallelization of individual fitness calculations
- threads (optional)
- the number of threads which are used for parallelization
Getter and Setter
mode
flag which indicates whether parallelization is used (True) or not (False)
thredNum
the number of threads which are used by enabled parallelization
References
| [Holland1975] | Holland, J. H. (1975). Adaptation in natural and artificial systems. University of Michigan Press, Ann Arbor. |
| [EOdev] | Keijzer, M., Merelo, J. J., Romero, G., & Schoenauer, M. (2002). Evolving objects: A general purpose evolutionary computation library. Artificial Evolution, 2310, pp. 829–888. |
| [Bolten2012] | Bolten, T. (2012) Genetische Algorithmen zur Auswahl und Gewichtung von Features in der Mustererkennung. Master thesis, Niederrhein University of Applied Sciences, Krefeld, Germany |